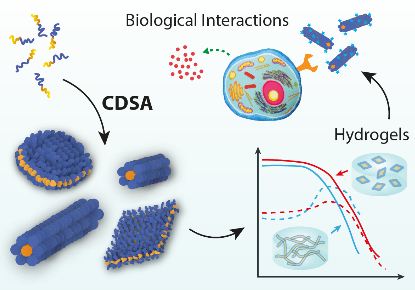
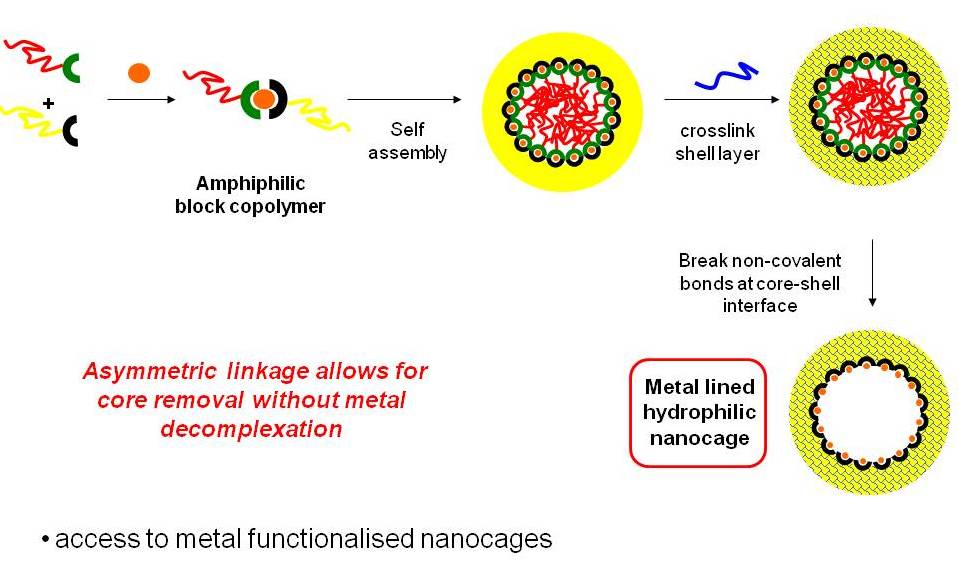
Biomimetic Nanotechnology
Nature routinely achieves feats of materials engineering that are currently very difficult for chemists to match. This is founded on precise control over the organisation of matter across multiple length scales, from the molecular level up to whole organisms. Our research seeks new ways of mimicking this advanced behaviour by taking motifs from biology – for example nucleic acid base pairing – and using them in synthetic systems. Exploiting the specificity of biological interactions allows us to design nanomaterials with unique, responsive behaviours, and to programme the synthesis of artificial sequence-defined polymers.
DNA-Templated Synthesis
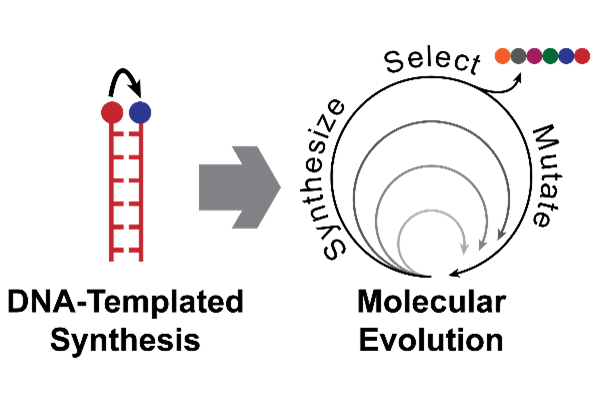
In our cells, DNA exists as a double helix – two strands held together by hydrogen bonds between complementary bases (A, T, G and C). These bonds are highly specific – A:T and G:C – and this can be exploited to programme DNA to adopt many different shapes and configurations at the nanoscale. It can also be exploited to control chemical reactions, by tethering reactive groups to the end of complementary strands of DNA: double helix formation brings these groups into close proximity, allowing a reaction to take place. This basic concept has applications in fields as diverse as sensing and catalysis. We are particularly interested in using it to mimic one of nature’s most impressive nanomachines: the ribosome. This nucleic acid/protein complex is responsible for translating genetic DNA into proteins – long chains of amino acids that fold into complex structures and perform all the jobs needed to sustain life. The ability to mimic this behaviour to make completely artificial, sequence-defined polymers would enable the development of truly advanced materials, sorely needed to address the many challenges currently facing humanity.
Evolution of DNA Templated Synthesis as a Tool for Materials Discovery, R. K. O’Reilly, A.J. Turberfield and T.R. Wilks, Acc. Chem. Res., 2017, 50, 2496–2509, DOI: 10.1021/acs.accounts.7b00280.
An autonomous molecular assembler for programmable chemical synthesis, Wenjing Meng, R. A. Muscat, M. L. McKee, P. J. Milnes, A. H. El-Sagheer, J. Bath, B. G. Davis, T. Brown, R. K. O’Reilly and A. J. Turberfield, Nature Chemistry, 2016, 8, 542–548, DOI: 10.1038/NChem.2495
Programmable one-pot multistep organic synthesis using DNA junctions, M. McKee, P. Milnes, J. Bath, E. Stulz, R.K. O’Reilly, A.J. Turberfield, J. Am. Chem. Soc. 2012, 134, 1446-1449. DOI: 10.1021/ja2101196
Biomolecule-Polymer Hybrids
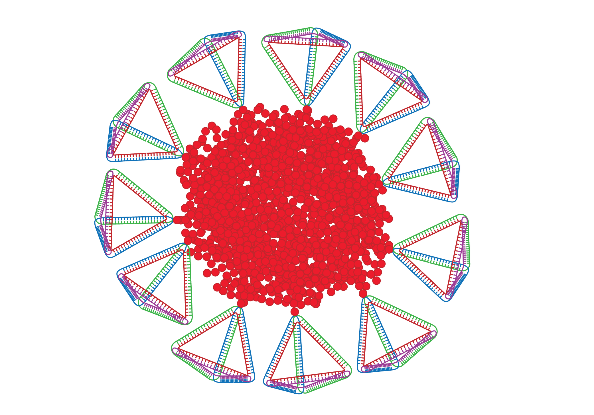
There is great interest in the preparation of biological-synthetic hybrid materials which combine the specificity of biomolecules with the scalability and robustness of polymers. DNA and proteins in particular endow synthetic polymers with new functions such as recognition and catalysis. Polymers, meanwhile, give access to a wide range of stimuli-responsive behaviours inaccessible to natural materials. Our research looks at new ways to efficiently attach biomolecules to polymers, and the unique ways in which the properties of these hybrids can be exploited for a range of applications including drug delivery and fundamental polymer self-assembly.
Self-assembly of temperature-responsive protein-polymer bioconjugates, D. Moatsou, J. Li, A. Ranji, A. Pitto-Barry, I. Ntai, M.C. Jewett, R.K. O’Reilly, Bioconjugate Chem, 2015, 26, 1890-1899. DOI: 10.1021/acs.bioconjchem.5b00264
Construction of DNA–polymer hybrids using intercalation interactions, T. Wilks, A. Pitto-Barry, N. Kirby, E. Stulz, R.K. O’Reilly, Chem Commun, 2014, 50, 1338-1340, DOI:10.1039/C3CC48726A.
‘Giant Surfactants’ Created by the Fast and Efficient Functionalization of a DNA Tetrahedron with a Temperature-Responsive Polymer, T. Wilks, J. Bath, Jan de Vries, J. Raymond, A. Herrmann, A.J. Turberfield, R.K. O’Reilly, ACS Nano, 2013, 7, 8561-8572. DOI:10.1021/nn402642a
Biomimetic Polymers
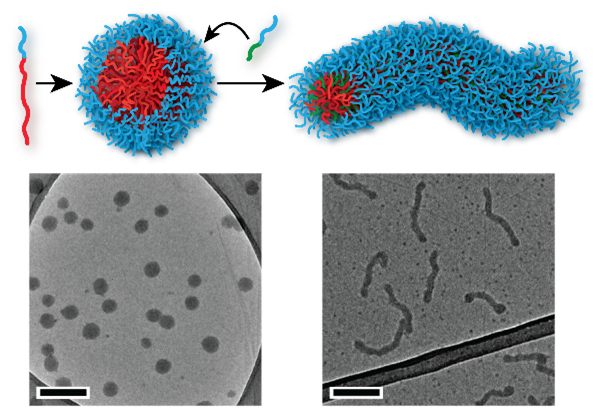
Proteins and DNA form the basis for all life on earth, by performing functions including data storage, catalysis and replication. Both are polymers, so it is reasonable to ask whether man-made polymers might be capable of similarly complex behaviour. One of the key features of natural polymers like proteins is that they have in-built recognition elements that interact and cause the molecule to fold into a complex structure, for example to create a pocket that holds a substrate in just the right position to promote a chemical reaction. In our research, we take these recognition elements and insert them into artificial polymers, with the objective of accessing complex self-assembly behaviour. Using this approach, we have managed to gain unprecedented control over polymer nanoparticle formation, with potential applications in medicine and beyond.
Anisotropic polymer nanoparticles with controlled dimensions from the morphological transformation of isotropic seeds, Z. Hua, J. R. Jones, M. Thomas, M. C. Arno, A. Souslov, T. R. Wilks & R. K. O’Reilly, Nat. Commun., 2019, 10, 5406-5413 , DOI: 10.1038/s41467-019-13263-6
Entrapment and Rigidification of Adenine by a Photo-Cross-Linked Thymine Network Leads to Fluorescent Polymer Nanoparticles, Z. Hua, T. R. Wilks, R. Keogh, G. Herwig, V. G. Stavros, and R. K. O’Reilly, Chemistry of Materials, 2018, 30, 4, 1408-1416, DOI: 10.1021/acs.chemmater.7b05206
Reversibly Manipulating the Surface Chemistry of Polymeric Nanostructures via a “Grafting To” Approach Mediated by Nucleobase Interactions, Z. Hua, R. Keogh, Z. Li, T. R. Wilks, G. Chen , and R. K. O’Reilly, Macromolecules, 2017, 50, 3662–3670, DOI: 10.1021/acs.macromol.7b00286